Integrative Taxonomy
Principal investigators: Rolando Gittens & Jose R. Loaiza
Collaborators:Luis Fernando de Leon, Alejandro Almanza, Luis Mejia, Fernando Merchan, Kelly L. Bennett, Javier Sanchez Galan, Jose Rovira & Matthew J. Miller
Background
Matrix-assisted laser desorption/ionization (MALDI) time-of-flight mass spectrometry is an analytical technique that allows for the sensitive and accurate detection of complex molecules such as proteins, peptides, lipids and nucleic acids. MALDI TOF MS has been used to discriminate among species of mosquitoes (Culicidae – Anopheles), fleas (Pulicidae – Ctenocephalidae), biting midges (Ceratopogonidae – Culicoides), sandflies (Psychodidae – Phlebotomus, Lutzomyia) and ticks (Ixodidae – Rhipicephalus). A key finding from these studies was that protein spectra obtained from body sections or whole specimens were similar among individuals of the same morphological species but differed noticeably across discrete morphological taxa. Therefore, MALDI protein spectra can be used as a reasonable tool to delimit species boundaries in arthropods that are vectors for pathogens that cause disease to humans.
Most existing applications of MALDI to identify arthropod disease vectors have focused on species-poor assemblages from Europe, but this technique is relatively less tested in the new world tropics. Properly identifying disease vectors such as ticks and mosquitoes in a highly diverse Neotropical country like Panama is a critical first step to predict and manage tick-borne zoonotic pathogens such as Rickettsia and arboviruses (e.g., arthropod-borne viruses) in Panama.
Findings & developments
- We developed an innovative and rapid method based on mass spectrometry to identify arthropod eggs, larvae/nymphs and adult body parts, which can be used for vector surveillance and identification by the health authorities in residential areas, ports of entry and forested environments of Panama.
- We generated a methodology that allows the identification of field-collected mosquitos from the Anophelesgenus without prior establishment of a reference library of well-curated lab-reared mosquitoes. This approach can be easily adapted and applied more broadly to other tropical regions of the world where Anophelesspecies diversity is high and morphological species complexes do exist.
- We established a procedure for the identification of mosquito eggs using mass spectrometry, thus allowing for the rapid evaluation of the presence of Ae. aegyptiand Ae. albopictus in garages trading used tires along highways, affording time and cost advantages over traditional vector surveillance.
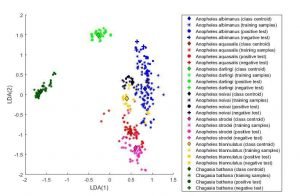
- The supervised nature Linear discriminant analysis—LDA classification algorithm is able to identify field-collected Anopheles species without the need of a reference library of species-specific protein spectra, and with higher resolution and discriminant power than unsupervised Principal component analysis—PCA.
Relevant publications
- Loaiza JR., † Scott ME, Bermingham E, Rovira J, Sanjur O, Conn JE. 2009. Short report: Anopheles darlingi in Panama. American Journal of Tropical Medicine and Hygiene 81: 23-26.
- Loaiza JR., † Scott ME, Bermingham E, Sanjur OI, Rovira Linton YM, et al. 2013. Novel genetic diversity within An. punctimacula s.l.: Phylogenetic discrepancy between the Barcode cytochrome c oxidase I gene and the rDNA second internal transcribed spacer. Acta Tropica 128(1):61-9.
- Loaiza JR., † Alejandro Almanza, Juan C. Rojas, Luis Mejia, Norma D. Cervantes, Javier Sanchez Galan, et al. 2019. Application of matrix-assisted laser desorption ionization mass spectrometry to identify species of the Neotropical malaria vector Anopheles. Malaria Journal, 18:95.